Projects
Highlighted
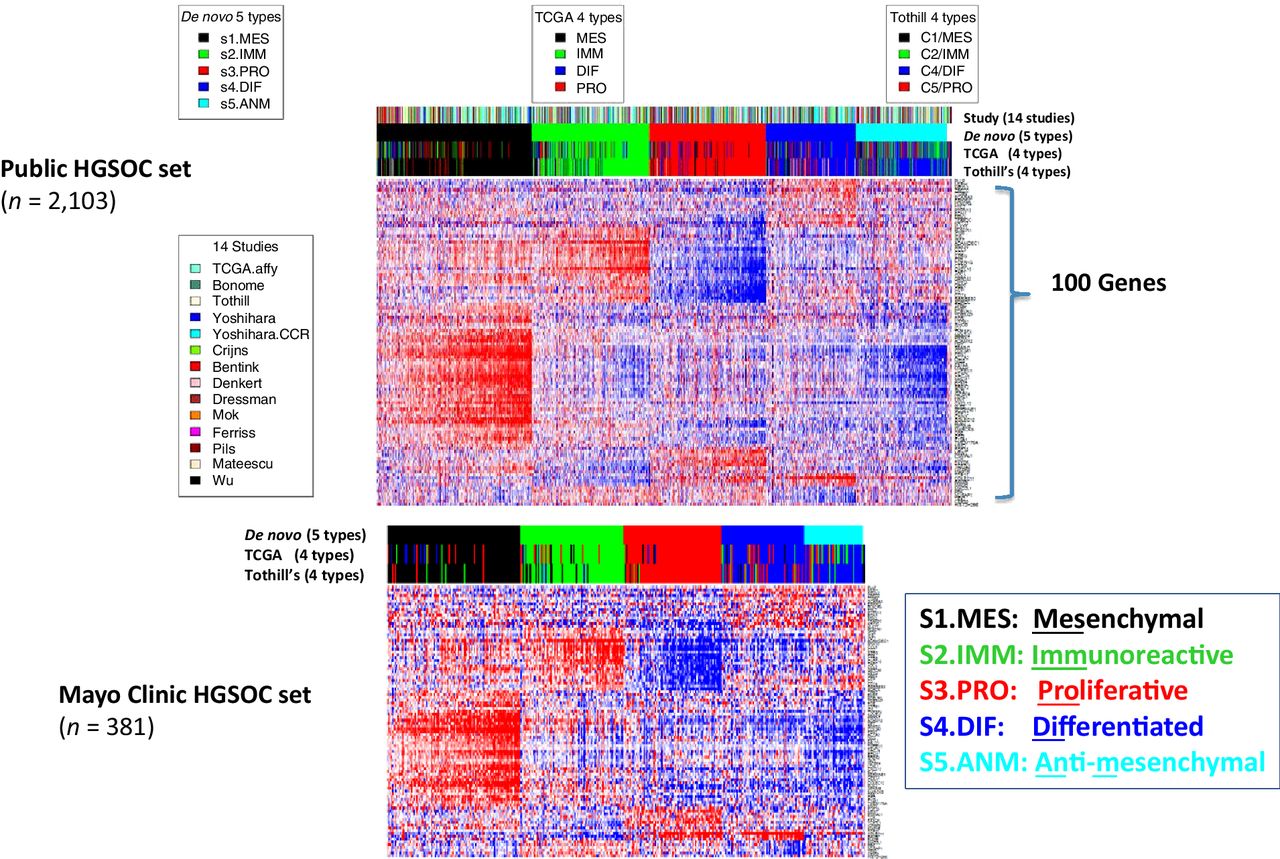
Despite growing knowledge about transcriptional profile-based molecular subtyping of high-grade serous ovarian cancer (HGSOC), molecular subtypes are not in current clinical use. Here we assess whether molecular subtyping identifies biological features of tumors that correlate with survival and surgical outcomes. In this study, five molecular subtypes were identified in a large public cohort (over 2,000 HGSOC patients) and validated in Mayo Clinic patients (381 patients). Molecular subtypes were significantly associated with surgical outcome and overall survival in both patient collections. The mesenchymal subtype of ovarian tumors generally showed the least favorable surgical outcome, with the lowest rate of no macroscopic disease (RD0) after primary surgeries. This result might suggest molecular subtyping should be clinically evaluated for future utility in guiding neoadjuvant treatment decisions for women with ovarian cancer.
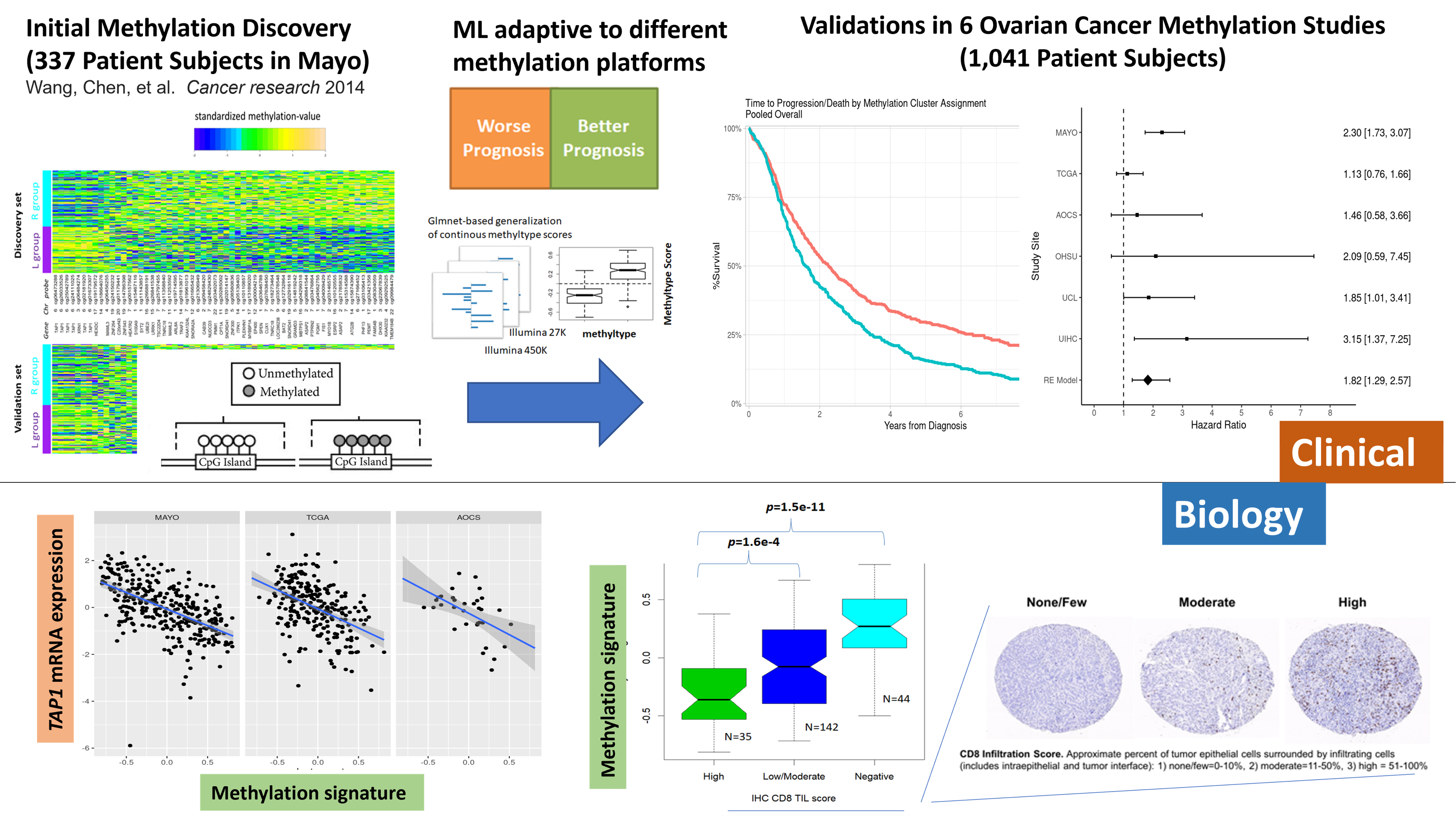
This study examined high-grade serous carcinoma (HGSC) of the tubo-ovarian cancers on DNA methylation as a prognostic factor. The study analyzed genome-wide DNA methylation in 1,040 HGSC samples, including 325 previously reported samples, to develop a quantitative methylation signature. This signature was found to be associated with a shorter time to recurrence, independent of clinical factors, and remained significant even after adjusting for gene expression molecular subtype and TAP1 expression. The methylation signature was inversely related to CD8+ tumor-infiltrating lymphocytes (TIL) levels and TAP1 expression and was associated with gene expression molecular subtype. The results suggest that DNA methylation could be a valuable prognostic tool in HGSC.
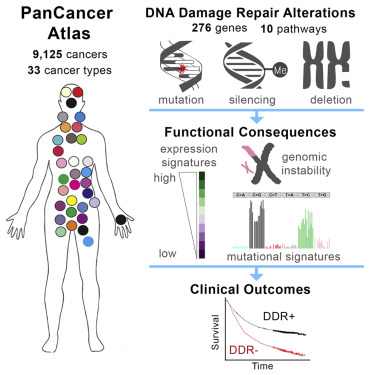
DNA damage repair (DDR) pathways modulate cancer risk, progression, and therapeutic response. We systematically analyzed somatic alterations to provide a comprehensive view of DDR deficiency across 33 cancer types. Mutations with accompanying loss of heterozygosity were observed in over 1/3 of DDR genes, including TP53 and BRCA1/2. Other prevalent alterations included epigenetic silencing of the direct repair genes EXO5, MGMT, and ALKBH3 in ∼20% of samples. Homologous recombination deficiency (HRD) was present at varying frequency in many cancer types, most notably ovarian cancer. However, in contrast to ovarian cancer, HRD was associated with worse outcomes in several other cancers. Protein structure-based analyses allowed us to predict functional consequences of rare, recurrent DDR mutations. A new machine-learning-based classifier developed from gene expression data allowed us to identify alterations that phenocopy deleterious TP53 mutations. These frequent DDR gene alterations in many human cancers have functional consequences that may determine cancer progression and guide therapy..
Datasets, Tools and Pipelines

A high quality ovarian tissue micro-array (TMA) dataset consists of multi-modality of images, with corresponding tabular data quantifying gene profiles of an ovarian cancer cohort.

A cell clustering pipeline developed for MxIF images. Developed with Nextflow, this tool is compatible and scalable on any POSIX system (Linux, OS X, etc).
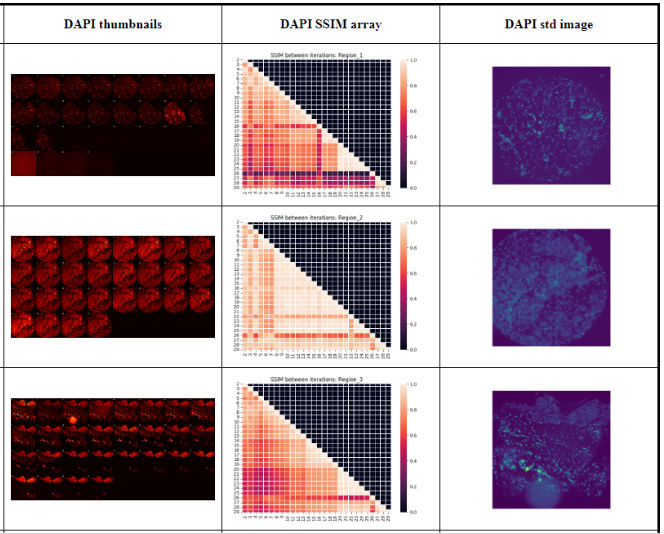
This tool has been developed to generate metrics and report the image quality coming from he Cytiva/Leica INCEll MxIF Microscope.
